INDI Library v2.0.7 is Released (01 Apr 2024)
Bi-monthly release with minor bug fixes and improvements
New KStars/Ekos Module: Analyze
- Ferrante Enriques
-

- Offline
- Elite Member
-

- Posts: 249
- Thank you received: 62
Replied by Ferrante Enriques on topic New KStars/Ekos Module: Analyze
I used Analyze on a set of images taken last night and it helped a lot to choose which were good or not before downloading from the observatory.
To me it would also be useful if images that don't match some criteria (e.g. HFR>1.5, Eccentricity>0.6) could be discarded /deleted before saving.
Ferrante
Please Log in or Create an account to join the conversation.
Replied by Jim on topic New KStars/Ekos Module: Analyze
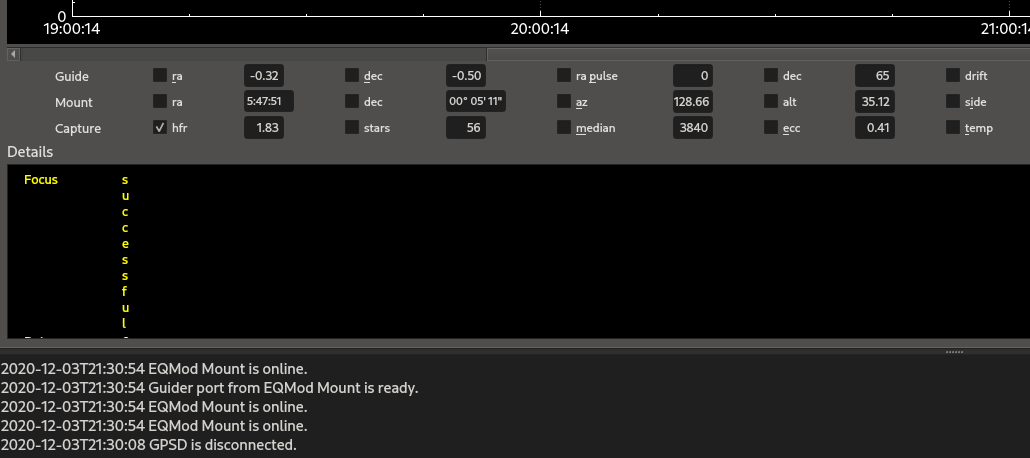
First, when I click on a "focus" segment or an captured image segment in the top section, I get these logs:
[2020-12-03T22:48:40.694 CST WARN ][ default] - QString::arg: Argument missing: <tr><td style="color:yellow">HFR</td><td colspan="2">1.80</td></tr>,
[2020-12-03T22:48:40.695 CST WARN ][ default] - QString::arg: Argument missing: <tr><td style="color:yellow">Iterations</td><td colspan="2">18</td></tr>,
[2020-12-03T22:48:40.695 CST WARN ][ default] - QString::arg: Argument missing: <tr><td style="color:yellow">Filter</td><td colspan="2">Red</td></tr>,
[2020-12-03T22:48:40.695 CST WARN ][ default] - QString::arg: Argument missing: <tr><td style="color:yellow">Temperature</td><td colspan="2">6.0</td></tr>, Second, when I click on a focus segment in the top section, the details are vertical with 1 letter width (see attached image).
Thanks
Jim
Please Log in or Create an account to join the conversation.
- Hy Murveit
-
 Topic Author
Topic Author
- Away
- Administrator
-

- Posts: 1222
- Thank you received: 565
Replied by Hy Murveit on topic New KStars/Ekos Module: Analyze
Can you say what your locale is? Can you test in other locales and see if it works? (Sorry, I'm new to this and don't really
know how to test with different locales). Would be happy to try and fix it if we knew more.
Hy
Please Log in or Create an account to join the conversation.
- Scott Denning
-

- Offline
- Elite Member
-

- Posts: 300
- Thank you received: 57
Replied by Scott Denning on topic New KStars/Ekos Module: Analyze
Would you consider turning off the guide panel if there's no guider set up, and then expanding gate vertical scale of the top two panels to fill the window?
I'm imaging unguided and my most important metric is eccentricity. There's so little vertical scale between 0 and 1 on the Statistics panel that it's hard to see what's going on.
Thanks again,
Scott
Please Log in or Create an account to join the conversation.
- Hy Murveit
-
 Topic Author
Topic Author
- Away
- Administrator
-

- Posts: 1222
- Thank you received: 565
Replied by Hy Murveit on topic New KStars/Ekos Module: Analyze
I'm definitely struggling with how to plot all the different possible graphs there.
I'm not sure what you mean. When you say "turning off the guide panel", do you mean the bottom two displays (labelled Details)?
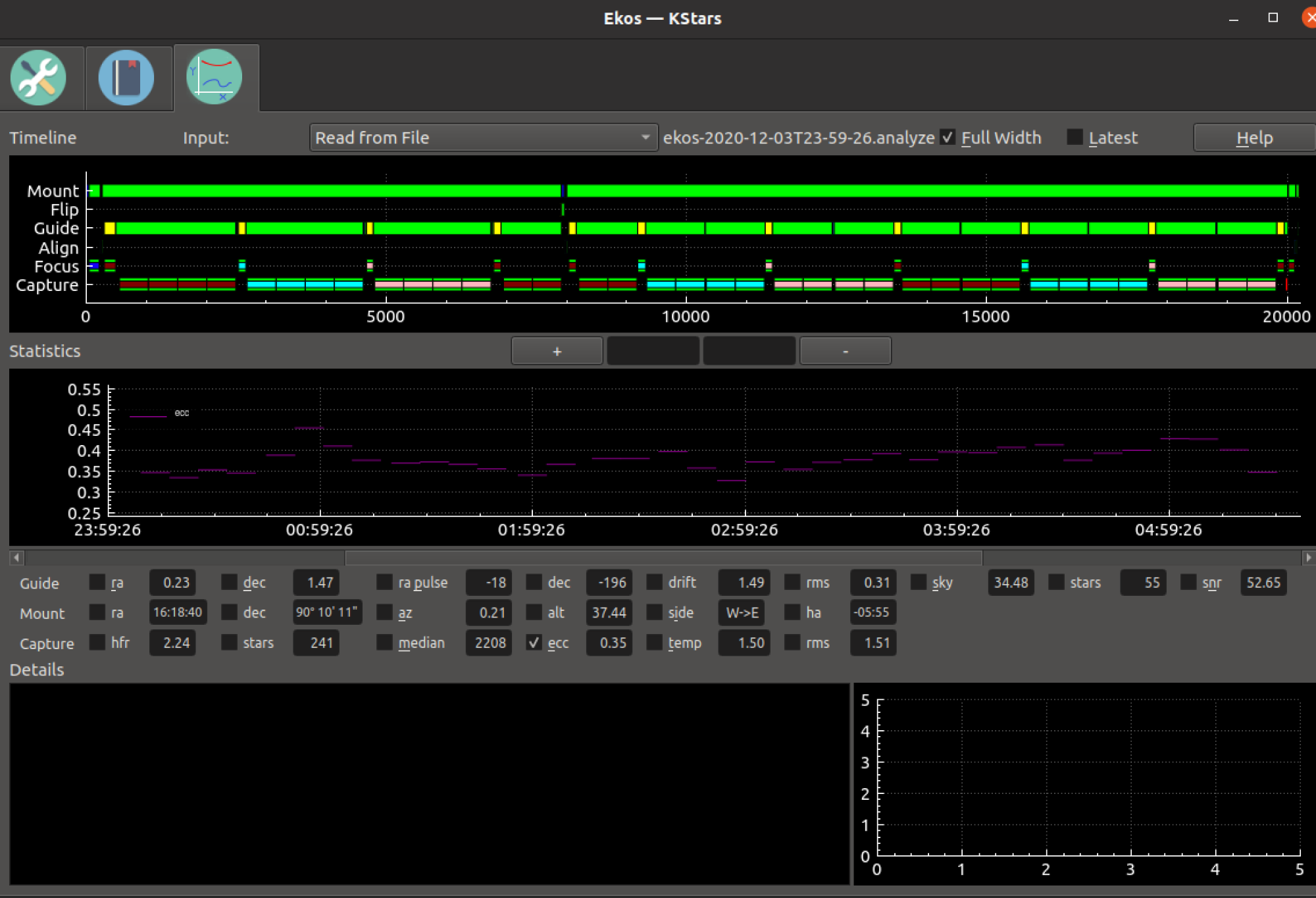
In any event, as a work around, until we figure something out (keep the suggestions coming), did you know that
you can put your mouse over a spot on the graph (e.g. right on some eccentricity value) and use the scroll where to zoom in to that spot.
You can then move your mouse over to the left to the y-axis, and drag the y-axis up and down.
If you use this technique, and turn off most other things, you can get a graph of eccentricity.
Here's a screenshot of me doing that.
I also want to warn you, that if you have quick HFR turned on, eccentricity is just calculated from the middle 25% of the image.
You can of course turn that off, but it may be computationally expensive on, e.g. a Raspberry Pi.
Hy
Please Log in or Create an account to join the conversation.
- Scott Denning
-

- Offline
- Elite Member
-

- Posts: 300
- Thank you received: 57
Replied by Scott Denning on topic New KStars/Ekos Module: Analyze
That being said, what is the point of those two bottom panels marked "Details" if there's no guider? Seems like a lot of screen space for two blank panels.
For that matter, the Guide, Mount, and Capture rows of tick boxes could be collapsed under a pull-down or moved to a Settings panel to allow more space for Statistics.
The guide data is important, and it's certainly great to see it here in the context of the rest of an imaging session, but it would be excellent to toggle it off if there's no guider present at all.
Thanks again,
Scott
Please Log in or Create an account to join the conversation.
Replied by Jim on topic New KStars/Ekos Module: Analyze
Interestingly, other sections display correctly, it is only the focus details, as if it were trying to print into a narrow column in that field. I am using Qt 5.15-1 if that helps. I don't know much about LOCALES either, and have not changed them from the default system install, but will look into it.
Jim
Please Log in or Create an account to join the conversation.
- Hy Murveit
-
 Topic Author
Topic Author
- Away
- Administrator
-

- Posts: 1222
- Thank you received: 565
Replied by Hy Murveit on topic New KStars/Ekos Module: Analyze
FWIW,
- The details graph on the right side also gives you an autofocus graph if you click the timeline section for an autofocus, and the left text section gives you information on any timeline section you click.
- I use the numbers coming from the checkbox section more than I use the graphs. E.g. I'll click on the graph section to find a time and look at various numbers. You can even pan around in the graph and the numbers should update with your mouse.
It may also be possible to pop-out a graph of interest.
Food for thought.
Hy
Please Log in or Create an account to join the conversation.
- Scott Denning
-

- Offline
- Elite Member
-

- Posts: 300
- Thank you received: 57
Replied by Scott Denning on topic New KStars/Ekos Module: Analyze
Please Log in or Create an account to join the conversation.
Replied by Palmito on topic New KStars/Ekos Module: Analyze
But Man!!! Thank you so much, your module is so nice!
I Love absolutely everything about it, what a sweet addition to ekos.
--edit: in a way bad weather was perfect to test your module, as I could see drops in guiding SNR curves, assess quality of my gathering and evaluate situation....
Please Log in or Create an account to join the conversation.
Replied by Bart on topic New KStars/Ekos Module: Analyze
-first of all- I use it a lot, it's an excellent overview and it really helps to better understand the overall session.
In the scheduler, I plan a few targets. The images from them can be opened by double clicking on the 'image trace' in the analyze tab.
But only for the first target.
When clicking the image trace for the next target, a popup shows noticing the image can't be found.
Can this be 'fixed'?
I use other names for my filters, so hereby I'm adding more names for the same filter in your code (see below).
I put it in <strong>bold</strong> font what I added. Feel free to add what you like (or nothing)
Cheers!bool filterStripeBrush(const QString &filter, QBrush *brush)
{
if (!filter.compare("red", Qt::CaseInsensitive) ||
!filter.compare("r", Qt::CaseInsensitive))
{
*brush = QBrush(Qt::red, Qt::SolidPattern);
return true;
}
else if (!filter.compare("green", Qt::CaseInsensitive) ||
!filter.compare("g", Qt::CaseInsensitive))
{
*brush = QBrush(Qt::green, Qt::SolidPattern);
return true;
}
else if (!filter.compare("blue", Qt::CaseInsensitive) ||
!filter.compare("b", Qt::CaseInsensitive))
{
*brush = QBrush(Qt::blue, Qt::SolidPattern);
return true;
}
else if (!filter.compare("ha", Qt::CaseInsensitive) ||
!filter.compare("h", Qt::CaseInsensitive) ||
<strong>!filter.compare("h-a", Qt::CaseInsensitive) ||
!filter.compare("h_a", Qt::CaseInsensitive) ||</strong>
<strong>!filter.compare("h-alpha", Qt::CaseInsensitive) ||
!filter.compare("hydrogen", Qt::CaseInsensitive) ||
!filter.compare("hydrogen_alpha", Qt::CaseInsensitive) ||
!filter.compare("hydrogen-alpha", Qt::CaseInsensitive) ||</strong>
!filter.compare("h_alpha", Qt::CaseInsensitive) ||
!filter.compare("halpha", Qt::CaseInsensitive))
{
*brush = QBrush(Qt::darkRed, Qt::SolidPattern);
return true;
}
else if (!filter.compare("oiii", Qt::CaseInsensitive) ||
<strong>!filter.compare("oxygen", Qt::CaseInsensitive) ||
!filter.compare("oxygen_3", Qt::CaseInsensitive) ||
!filter.compare("oxygen-3", Qt::CaseInsensitive) ||
!filter.compare("oxygen_iii", Qt::CaseInsensitive) ||
!filter.compare("oxygen-iii", Qt::CaseInsensitive) ||
!filter.compare("o_iii", Qt::CaseInsensitive) ||
!filter.compare("o-iii", Qt::CaseInsensitive) ||</strong>
<strong>!filter.compare("o_3", Qt::CaseInsensitive) ||
!filter.compare("o-3", Qt::CaseInsensitive) ||</strong>
!filter.compare("o3", Qt::CaseInsensitive))
{
*brush = QBrush(Qt::cyan, Qt::SolidPattern);
return true;
}
else if (!filter.compare("sii", Qt::CaseInsensitive) ||
<strong>!filter.compare("sulphur", Qt::CaseInsensitive) ||
!filter.compare("sulphur_2", Qt::CaseInsensitive) ||
!filter.compare("sulphur-2", Qt::CaseInsensitive) ||
!filter.compare("sulphur_ii", Qt::CaseInsensitive) ||
!filter.compare("sulphur-ii", Qt::CaseInsensitive) ||
!filter.compare("s_ii", Qt::CaseInsensitive) ||
!filter.compare("s-ii", Qt::CaseInsensitive) ||</strong>
<strong>!filter.compare("s_2", Qt::CaseInsensitive) ||</strong>
<strong>!filter.compare("s-2", Qt::CaseInsensitive) ||</strong>
!filter.compare("s2", Qt::CaseInsensitive))
{
// Pink.
*brush = QBrush(QColor(255, 182, 193), Qt::SolidPattern);
return true;
}
else if (!filter.compare("lpr", Qt::CaseInsensitive) ||
!filter.compare("L", Qt::CaseInsensitive) ||
<strong>!filter.compare("UV-IR cut", Qt::CaseInsensitive) ||</strong>
<strong>!filter.compare("UV-IR", Qt::CaseInsensitive) ||</strong>
<strong>!filter.compare("white", Qt::CaseInsensitive) ||</strong>
<strong>!filter.compare("monochrome", Qt::CaseInsensitive) ||</strong>
<strong>!filter.compare("broadband", Qt::CaseInsensitive) ||</strong>
<strong>!filter.compare("clear", Qt::CaseInsensitive) ||</strong>
<strong>!filter.compare("focus", Qt::CaseInsensitive) ||</strong>
!filter.compare("luminance", Qt::CaseInsensitive) ||
!filter.compare("lum", Qt::CaseInsensitive) ||
!filter.compare("lps", Qt::CaseInsensitive) ||
!filter.compare("cls", Qt::CaseInsensitive))
{
*brush = QBrush(Qt::white, Qt::SolidPattern);
return true;
}
return false;
}
Please Log in or Create an account to join the conversation.
- Hy Murveit
-
 Topic Author
Topic Author
- Away
- Administrator
-

- Posts: 1222
- Thank you received: 565
Replied by Hy Murveit on topic New KStars/Ekos Module: Analyze
Re the new filter synonyms: great suggestions. I just put in a merge request to have those added, pending review by Jasem.
See invent.kde.org/education/kstars/-/merge_requests/318
Re the bug you report:
As I understand, are you saying that when you double click on a capture in the timeline, and you do this for the 2nd scheduler job in an evening, the image doesn't display? I tried to reproduce with the simulator but it worked ok for me. Can you please single click on one of the captures in the timeline that doesn't display, and look at the filename shown in the "Details" table, and make sure that file exists on disk? Is there something odd about the filename? Can you reproduce with the simulator? Any clues would be helpful. FWIW, someone did make modifications to the file naming in Ekos recently, so I suppose this could have caused issues here.
Thanks,
Hy
Please Log in or Create an account to join the conversation.